2/03/2006
Dead Man's Sperm is not Legal Father
The Tokyo High Court refused Wednesday to recognize a girl conceived using sperm taken from a man before he died of illness as his, upholding a lower court decision.
The way the girl came to be born "significantly deviates" from natural reproduction and there is no common social recognition accepting reproduction assisted by in vitro medical technology, Presiding Judge Kimio Miyazaki said in handing down the ruling.
In 2001, according to the ruling, the man had his sperm taken on five occasions and it was kept frozen at a medical institution. By the time he died the next year, three in vitro fertilization attempts had failed. The woman became pregnant on the fourth attempt, after his death.
"Reproduction after the donor's death leads to creating a fatherless child, and such an act is unacceptable in view of the child's welfare," he added.
The lower court judge had said the donor's consent for his sperm to be used for in-vitro fertilization must be confirmed each time an attempt is made and it cannot be construed that the man's consent was valid after his death.
LEAD: Court nixes appeal to recognize girl conceived with dead man's sperm
The way the girl came to be born "significantly deviates" from natural reproduction and there is no common social recognition accepting reproduction assisted by in vitro medical technology, Presiding Judge Kimio Miyazaki said in handing down the ruling.
In 2001, according to the ruling, the man had his sperm taken on five occasions and it was kept frozen at a medical institution. By the time he died the next year, three in vitro fertilization attempts had failed. The woman became pregnant on the fourth attempt, after his death.
"Reproduction after the donor's death leads to creating a fatherless child, and such an act is unacceptable in view of the child's welfare," he added.
The lower court judge had said the donor's consent for his sperm to be used for in-vitro fertilization must be confirmed each time an attempt is made and it cannot be construed that the man's consent was valid after his death.
LEAD: Court nixes appeal to recognize girl conceived with dead man's sperm
Descendants of Abraham Lincoln Reveal History
Researchers at Johns Hopkins and the University of Minnesota have discovered a gene mutation in the descendants of Abraham Lincoln’s grandparents that suggests the Civil War president himself might have also suffered from a disease that destroys nerve cells in the cerebellum-- the part of the brain that controls movement. A report on this discovery will appear in the February print issue of Nature Genetics.

The present discovery in Lincoln’s descendants of the gene that causes a movement disorder called spinocerebellar ataxia type 5 (SCA5.
The joint finding of the SCA5 mutation comes over a decade after initial speculation that Lincoln might have suffered from Marfan disease. People with this inherited disorder are often tall and thin and can commonly have slender, tapering fingers.
The researchers discovered that SCA5 is caused by a mutation of the β-III spectrin gene SPTBN2, which disrupts the ability of certain nerves in the cerebellum to respond normally to incoming chemical signals.
The mutation was found in 90 of 299 of Lincoln's descendants showing symptoms of the disease and 35 who did not have symptoms.
DISCOVERY OF MUTATION IN BRAIN CELLS OF DESCENDANTS OF ABRAHAM LINCOLN SUGGEST THE PRESIDENT SUFFERED FROM MOVEMENT DISORDER

The present discovery in Lincoln’s descendants of the gene that causes a movement disorder called spinocerebellar ataxia type 5 (SCA5.
The joint finding of the SCA5 mutation comes over a decade after initial speculation that Lincoln might have suffered from Marfan disease. People with this inherited disorder are often tall and thin and can commonly have slender, tapering fingers.
The researchers discovered that SCA5 is caused by a mutation of the β-III spectrin gene SPTBN2, which disrupts the ability of certain nerves in the cerebellum to respond normally to incoming chemical signals.
The mutation was found in 90 of 299 of Lincoln's descendants showing symptoms of the disease and 35 who did not have symptoms.
DISCOVERY OF MUTATION IN BRAIN CELLS OF DESCENDANTS OF ABRAHAM LINCOLN SUGGEST THE PRESIDENT SUFFERED FROM MOVEMENT DISORDER
Gene variation increases SIDS risk in African Americans
About five percent of deaths from SIDS (sudden infant death syndrome) in African Americans can be traced to defects in one gene a research team based at the University of Chicago reports in the February 2006, issue of the Journal of Clinical Investigation.
A specific mutation, called Y1103, in a gene called SCN5A is associated with a dramatic, 24-fold increased risk of sudden infant death syndrome (SIDS) in African American infants. The authors show that this mutant protein, when exposed to acidic conditions (which can be caused by low blood oxygen levels when infants are placed in the face-down or "prone" sleeping position), malfunctions in a way that has been previously shown to trigger an irregular heartbeat in adults.
But SIDS is not purely genetic; it appears to require multiple "hits," some from altered genes and some from the environment.
"The hope," said Steven Goldstein, MD, PhD, professor and chairman of pediatrics at the University of Chicago and director of the study, "is that findings like this may one day allow us to intervene. We might screen to identify children at high risk and teach parents how to lessen the likelihood of secondary challenges. We have already begun to evaluate drugs that may mitigate the risk."
University of Chicago Hospitals: Gene variation increases SIDS risk in African Americans
A specific mutation, called Y1103, in a gene called SCN5A is associated with a dramatic, 24-fold increased risk of sudden infant death syndrome (SIDS) in African American infants. The authors show that this mutant protein, when exposed to acidic conditions (which can be caused by low blood oxygen levels when infants are placed in the face-down or "prone" sleeping position), malfunctions in a way that has been previously shown to trigger an irregular heartbeat in adults.
But SIDS is not purely genetic; it appears to require multiple "hits," some from altered genes and some from the environment.
"The hope," said Steven Goldstein, MD, PhD, professor and chairman of pediatrics at the University of Chicago and director of the study, "is that findings like this may one day allow us to intervene. We might screen to identify children at high risk and teach parents how to lessen the likelihood of secondary challenges. We have already begun to evaluate drugs that may mitigate the risk."
University of Chicago Hospitals: Gene variation increases SIDS risk in African Americans
2/01/2006
Gene Mutation and Parkinson's Disease
January 26, 2006 — (BRONX, NY) — Researchers at the Albert Einstein College of Medicine of Yeshiva University and its Manhattan hospital affiliate, Beth Israel Medical Center, have found that a specific mutation in a single gene is a major cause of Parkinson’s disease among Ashkenazi (Eastern European) Jews. The report will appear in the January 26 issue of The New England Journal of Medicine.
“Like the discovery of the BRCA1 and BRCA2 gene mutations for breast cancer, this finding will directly affect the way Parkinson’s disease is diagnosed in Ashkenazi Jews," says Dr. Susan B. Bressman, senior investigator of the report, "It also emphasizes the benefit of focusing genetic studies in a specific ethnic group, even with regard to a disease not thought to be primarily genetic in origin."
"Our finding should bring genetic counseling for Parkinson’s disease to the forefront along with genetic testing for early detection of Parkinson’s disease." adds study co-author Dr. Laurie J. Ozelius, Associate Professor of Molecular Genetics at Einstein.
The researchers focused on a gene called LRRK2, which is mutated in about 1% of late-onset non-familial cases of Parkinson’s disease in those patients who are primarily of European ancestry.
In addition to Ashkenazi Jews, the researchers note that a group of North Africans of Arab descent have been found to have a high frequency of this same gene mutation as a cause of Parkinson’s disease. The two groups appear to share the same origin or founder, suggesting a probable Middle Eastern origin for this mutation.
AECOM: News Releases
“Like the discovery of the BRCA1 and BRCA2 gene mutations for breast cancer, this finding will directly affect the way Parkinson’s disease is diagnosed in Ashkenazi Jews," says Dr. Susan B. Bressman, senior investigator of the report, "It also emphasizes the benefit of focusing genetic studies in a specific ethnic group, even with regard to a disease not thought to be primarily genetic in origin."
"Our finding should bring genetic counseling for Parkinson’s disease to the forefront along with genetic testing for early detection of Parkinson’s disease." adds study co-author Dr. Laurie J. Ozelius, Associate Professor of Molecular Genetics at Einstein.
The researchers focused on a gene called LRRK2, which is mutated in about 1% of late-onset non-familial cases of Parkinson’s disease in those patients who are primarily of European ancestry.
In addition to Ashkenazi Jews, the researchers note that a group of North Africans of Arab descent have been found to have a high frequency of this same gene mutation as a cause of Parkinson’s disease. The two groups appear to share the same origin or founder, suggesting a probable Middle Eastern origin for this mutation.
AECOM: News Releases
Genetic Variations Implicated In Disease
Sequence differences in less than 0.2% of the 3-billion-base human genome play a vital role in a bewildering variety of human disease. Today, researchers from the Wellcome Trust Sanger Institute and the Cambridge University's Cambridge Institute for Medical Research, together with international colleagues report in PLoS Genetics their detailed maps of differences implicated in disease as well as genes that are unchanged in recent human history.
The Major Histocompatibility Complex (MHC) consists of hundreds of genes on human chromosome 6 that are important in most autoimmune conditions, when our biological defences turn on our own systems.
The MHC has the major role in type 1 diabetes and rheumatoid arthritis. The MHC is also pivotal in response to infection, including malaria and AIDS.
Genes in the MHC can differ dramatically between people, and the differences among us affect medical events as diverse as tissue transplant rejection, arthritis, asthma and disease resistance. A detailed study of this region in different people will shed light on which genes are most important.
"Within the sea of over 20,000 sequence variations across the 4 million MHC bases, we found one island of stability," continued Dr Beck. "A region of 160,000 bases that is up to 200-fold less variant between chromosomes sharing part of the same HLA type, suggesting these individuals most likely shared a common ancestor as recently as 50,000 years ago."
The study further described over 300 amino acid changing variants in gene sequences. These variants are strong candidates for functional studies to understand the role of variation in MHC-associated disease.
Autoimmune disease affects about 3 million people in the UK. The three haplotypes studied here display different susceptibilities to diseases such as type 1 diabetes, myasthenia gravis and multiple sclerosis.
Haplotypes are combinations of gene and sequence variants that tend to occur together in an individual genome. This may be purely fortuitous, or it may reflect selection of given combinations (they have been successful in the past), or it may reflect a population bottleneck, where only a few, perhaps similar, genomes have contributed to the further population growth.
The MHC is among the most gene-dense regions of the human genome and the most variable. Over evolutionary time, the MHC has been driven to become the most variable region of our genome.
The MHC Haplotype Project is studying in fine detail the sequence of eight of the most common human haplotypes, selected for conferring protection against or susceptibility to common disease. The detailed analysis of the third of these eight is reported here and compared with the two previously described.
The COX haplotype has been associated with susceptibility to a wide range of diseases, including type 1 diabetes, systemic lupus erythematosus and myasthenia gravis.
The PGF haplotype provides protection against type 1 diabetes and predisposes to other diseases such as multiple sclerosis and systemic lupus erythematosus.
The QBL haplotype is positively associated with Graves' disease and type 1 diabetes.
ScienceDaily: Researchers Map Of Genetic Variations Implicated In Disease
The Major Histocompatibility Complex (MHC) consists of hundreds of genes on human chromosome 6 that are important in most autoimmune conditions, when our biological defences turn on our own systems.

The MHC has the major role in type 1 diabetes and rheumatoid arthritis. The MHC is also pivotal in response to infection, including malaria and AIDS.
Genes in the MHC can differ dramatically between people, and the differences among us affect medical events as diverse as tissue transplant rejection, arthritis, asthma and disease resistance. A detailed study of this region in different people will shed light on which genes are most important.
"Within the sea of over 20,000 sequence variations across the 4 million MHC bases, we found one island of stability," continued Dr Beck. "A region of 160,000 bases that is up to 200-fold less variant between chromosomes sharing part of the same HLA type, suggesting these individuals most likely shared a common ancestor as recently as 50,000 years ago."
The study further described over 300 amino acid changing variants in gene sequences. These variants are strong candidates for functional studies to understand the role of variation in MHC-associated disease.
Autoimmune disease affects about 3 million people in the UK. The three haplotypes studied here display different susceptibilities to diseases such as type 1 diabetes, myasthenia gravis and multiple sclerosis.
Haplotypes are combinations of gene and sequence variants that tend to occur together in an individual genome. This may be purely fortuitous, or it may reflect selection of given combinations (they have been successful in the past), or it may reflect a population bottleneck, where only a few, perhaps similar, genomes have contributed to the further population growth.
The MHC is among the most gene-dense regions of the human genome and the most variable. Over evolutionary time, the MHC has been driven to become the most variable region of our genome.
The MHC Haplotype Project is studying in fine detail the sequence of eight of the most common human haplotypes, selected for conferring protection against or susceptibility to common disease. The detailed analysis of the third of these eight is reported here and compared with the two previously described.
The COX haplotype has been associated with susceptibility to a wide range of diseases, including type 1 diabetes, systemic lupus erythematosus and myasthenia gravis.
The PGF haplotype provides protection against type 1 diabetes and predisposes to other diseases such as multiple sclerosis and systemic lupus erythematosus.
The QBL haplotype is positively associated with Graves' disease and type 1 diabetes.
ScienceDaily: Researchers Map Of Genetic Variations Implicated In Disease
DNA Key to Predicting Prostate and Renal Cancer
In a study to be published in the February issue of "The Journal of Urology," researchers from Emory University School of Medicine have determined that inheritance of "mitochondrial haplogroup U" is associated with increased risk of prostate and renal cancers.
John Petros, MD, associate professor of Urology at Emory, led the study with colleagues from Emory and the Mercer University School of Medicine. Statistics from the study show that 20 million people in the United States fall into the haplogroup U category and are therefore at an increased risk for prostate or renal cancer.
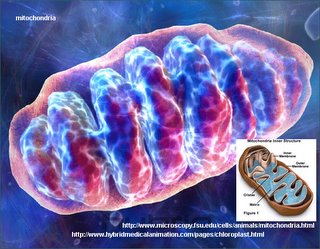
Mitochondrial DNA is passed down from the mother to her children, males and females. In this study, Dr. Petros and his colleagues researched the inheritance pattern of mitochondria in patients with cancer.
"The study found that inheritance of mitochondrial haplogroup U is associated with an approximately 2-fold increased risk of prostate cancer and 2.5-fold increased risk of renal cancer in white North American individuals," says Dr. Petros, "Mitochondrial haplogroup U is found in 9.35 percent of the white United States population, which means more than 20 million individuals are in this high risk group."
By comparing the mtDNA haplotype in patients with prostate and renal cancer to that in control groups, Petros has been able to determine an association between mitochondrial genotype and cancer risk.
A haplotype is a set of closely linked alleles, which are genes or variations in the sequence of genetic information on a segment of DNA. "Haplo" comes from the Greek word for "single."
Mitochondrial haplogroups have been associated with other diseases. For example, the H haplogroup has been linked to late onset Alzheimer's disease and the J haplogroup is connected to Liber's hereditary optic neuropathy.
Emory WHSC :: Press Releases :: Article Detail :: DNA Key to Predicting Prostate and Renal Cancer
John Petros, MD, associate professor of Urology at Emory, led the study with colleagues from Emory and the Mercer University School of Medicine. Statistics from the study show that 20 million people in the United States fall into the haplogroup U category and are therefore at an increased risk for prostate or renal cancer.

Mitochondrial DNA is passed down from the mother to her children, males and females. In this study, Dr. Petros and his colleagues researched the inheritance pattern of mitochondria in patients with cancer.
"The study found that inheritance of mitochondrial haplogroup U is associated with an approximately 2-fold increased risk of prostate cancer and 2.5-fold increased risk of renal cancer in white North American individuals," says Dr. Petros, "Mitochondrial haplogroup U is found in 9.35 percent of the white United States population, which means more than 20 million individuals are in this high risk group."
By comparing the mtDNA haplotype in patients with prostate and renal cancer to that in control groups, Petros has been able to determine an association between mitochondrial genotype and cancer risk.
A haplotype is a set of closely linked alleles, which are genes or variations in the sequence of genetic information on a segment of DNA. "Haplo" comes from the Greek word for "single."
Mitochondrial haplogroups have been associated with other diseases. For example, the H haplogroup has been linked to late onset Alzheimer's disease and the J haplogroup is connected to Liber's hereditary optic neuropathy.
Emory WHSC :: Press Releases :: Article Detail :: DNA Key to Predicting Prostate and Renal Cancer
Genetic Risk for Bipolar Disorder
A risk gene for bipolar disorder has been shown for the first time by a team including researchers from University of New South Wales and the Garvan Institute of Medical Research.
The research found those who have a particular form of this gene are twice as likely to develop the condition.
Bipolar disorder, which is also known as manic-depressive illness, affects two in every 100 people, with mood swings resulting in periods of mania and depression with normal behaviour between episodes.
"Apart from lithium, which a significant number of patients cannot tolerate, the currently available medicines are not specific for the condition," said Professor Mitchell. "The identification of this gene could allow for the development of targeted medicines."
tolerate, the currently available medicines are not specific for the condition," said Professor Mitchell. "The identification of this gene could allow for the development of targeted medicines."
"We used a number of families, unrelated patients and therapeutic drug models. Each of these led us to the same gene, called F-A-T 1," said UNSW Professor Peter Schofield, who led the team at the Garvan Institute of Medical Research and is now the Executive Director of the Prince of Wales Medical Research Institute (POWMRI).
Although the researchers have identified the gene, they are now trying to work out how it increases the risk of the disorder
UNSW: The University of New South Wales - Sydney Australia - News - Bipolar disorder: the genetic risk revealed
The research found those who have a particular form of this gene are twice as likely to develop the condition.
Bipolar disorder, which is also known as manic-depressive illness, affects two in every 100 people, with mood swings resulting in periods of mania and depression with normal behaviour between episodes.
"Apart from lithium, which a significant number of patients cannot
 tolerate, the currently available medicines are not specific for the condition," said Professor Mitchell. "The identification of this gene could allow for the development of targeted medicines."
tolerate, the currently available medicines are not specific for the condition," said Professor Mitchell. "The identification of this gene could allow for the development of targeted medicines.""We used a number of families, unrelated patients and therapeutic drug models. Each of these led us to the same gene, called F-A-T 1," said UNSW Professor Peter Schofield, who led the team at the Garvan Institute of Medical Research and is now the Executive Director of the Prince of Wales Medical Research Institute (POWMRI).
Although the researchers have identified the gene, they are now trying to work out how it increases the risk of the disorder
UNSW: The University of New South Wales - Sydney Australia - News - Bipolar disorder: the genetic risk revealed
Born to Dance
In a study published in the American journal, Public Library of Science Genetics, Psychology Prof. Richard P. Ebstein and his research associates have shown, through DNA examination, that dancers show consistent differences in two key genes from the general population. Ebstein is the head of the Hebrew University Psychology Department's Scheinfeld Center for Human Genetics in the Social Sciences.
This finding is not surprising, says Ebstein, in view of other studies of musicians and athletes, which also have shown genetic differences.

Ebstein and his colleagues found in an examination of 85 dancers and advanced dancing students in Israel variants of two genes that provide the code for the serotonin transporter and arginine vasopressin receptor 1a.
The serotonin transporter regulates the level of serotonin, a brain transmitter that contributes to spiritual experience, among many other behavioral traits. The vasopressin receptor has been shown in many animal studies to modulate social communication and affiliative bonding behaviors. Both are elements involved in the age-old human social expression of dancing.
The genetic evidence was corroborated by two questionnaires distributed by the researchers to the dancers. One is the Tellegen Absorption Scale (TAS), that correlates aspects of spirituality and altered states of consciousness, and the other is the Tridimensional Personality Questionnaire (TPQ), a measure of the need for social contact and openness to communication.
The genetic and questionnaire results of the dancers were compared with those of two other groups examined – athletes as well as those who were both non-dancers and non-athletes. (Athletes were chosen for comparison since they require a good deal of physical stamina like dancers.)
The dancer "type," says Ebstein, clearly demonstrates qualities that are not necessarily lacking but are not expressed as strongly in other people: a heightened sense of communication, often of a symbolic and ceremonial nature, and a strong spiritual personality trait.
Are dancers genetically different than the rest of us? Yes, says Hebrew University researcher
This finding is not surprising, says Ebstein, in view of other studies of musicians and athletes, which also have shown genetic differences.

Ebstein and his colleagues found in an examination of 85 dancers and advanced dancing students in Israel variants of two genes that provide the code for the serotonin transporter and arginine vasopressin receptor 1a.
The serotonin transporter regulates the level of serotonin, a brain transmitter that contributes to spiritual experience, among many other behavioral traits. The vasopressin receptor has been shown in many animal studies to modulate social communication and affiliative bonding behaviors. Both are elements involved in the age-old human social expression of dancing.
The genetic evidence was corroborated by two questionnaires distributed by the researchers to the dancers. One is the Tellegen Absorption Scale (TAS), that correlates aspects of spirituality and altered states of consciousness, and the other is the Tridimensional Personality Questionnaire (TPQ), a measure of the need for social contact and openness to communication.
The genetic and questionnaire results of the dancers were compared with those of two other groups examined – athletes as well as those who were both non-dancers and non-athletes. (Athletes were chosen for comparison since they require a good deal of physical stamina like dancers.)
The dancer "type," says Ebstein, clearly demonstrates qualities that are not necessarily lacking but are not expressed as strongly in other people: a heightened sense of communication, often of a symbolic and ceremonial nature, and a strong spiritual personality trait.
Are dancers genetically different than the rest of us? Yes, says Hebrew University researcher
Genes Controlled by DNA Snippets
"Most people don’t realize that genes make up a very small percentage of the human DNA code," said Joseph M. Miano, Ph.D., senior author of a Genome Research journal paper and associate professor within the Cardiovascular Research Institute at the University of Rochester Medical Center. “Genes are relatively straightforward compared to what lies ahead. We believe that the real genetic gymnastics, the real intelligence of our system, is controlled by tiny bits of genetic material that tell genes what to do."
Genes are the chains of deoxyribonucleic acids (DNA) that encode instructions for the building of proteins, the workhorses that make up the body’s organs and carry its signals.
The Human Genome Project, which first reported results in 2001, produced a near complete listing of the DNA sequences that make up all human genes (the genome). Key project findings included that human genetic material consists of about 3 billion base pairs, the “letters" that make up the genetic code. Researchers also concluded that genes, specific batches of code that direct protein construction, comprise just about 2 percent of all human DNA. A central question in genetics has become: what does the remaining 98 percent of human genetic material do?
Regulatory sequences are emerging as an important part of the non-gene majority of human genetic material, once thought of as “junk DNA."
In Miano’s study, the regulatory sequence under examination was the CArG box. The nucleotide building blocks of DNA chains may contain any one of four nucleobases: adenine (A), thymine (T), guanine (G) and cytosine (C). Any sequence of code starting with 2 Cs, followed by any combination of 6 As or Ts, and ending in 2 Gs is a CArG box.
According to Miano, there are 1,216 variations of CArG box that together occur approximately three million times throughout the human DNA blueprint.
CArG boxes exert their influence over genes because they are “shaped" to partner with several proteins within a genetic regulatory network. Throughout a human life, such networks are believed to “decide" the timing and location of all gene expression, the process through which genetic information is converted into templates for protein construction.
As part of this effort, researchers at the University of Rochester Medical Center scanned through the vast human DNA code to reveal for the first time 60 genes influenced by one such sequence.
Why is this important?
The Human Genome Project was just the beginning for understanding how genes affect the human body.
Study Finds 60 New Genes Controlled by DNA Snippet - URMC Press Room

Genes are the chains of deoxyribonucleic acids (DNA) that encode instructions for the building of proteins, the workhorses that make up the body’s organs and carry its signals.
The Human Genome Project, which first reported results in 2001, produced a near complete listing of the DNA sequences that make up all human genes (the genome). Key project findings included that human genetic material consists of about 3 billion base pairs, the “letters" that make up the genetic code. Researchers also concluded that genes, specific batches of code that direct protein construction, comprise just about 2 percent of all human DNA. A central question in genetics has become: what does the remaining 98 percent of human genetic material do?
Regulatory sequences are emerging as an important part of the non-gene majority of human genetic material, once thought of as “junk DNA."
In Miano’s study, the regulatory sequence under examination was the CArG box. The nucleotide building blocks of DNA chains may contain any one of four nucleobases: adenine (A), thymine (T), guanine (G) and cytosine (C). Any sequence of code starting with 2 Cs, followed by any combination of 6 As or Ts, and ending in 2 Gs is a CArG box.
According to Miano, there are 1,216 variations of CArG box that together occur approximately three million times throughout the human DNA blueprint.
CArG boxes exert their influence over genes because they are “shaped" to partner with several proteins within a genetic regulatory network. Throughout a human life, such networks are believed to “decide" the timing and location of all gene expression, the process through which genetic information is converted into templates for protein construction.
As part of this effort, researchers at the University of Rochester Medical Center scanned through the vast human DNA code to reveal for the first time 60 genes influenced by one such sequence.
Why is this important?
The Human Genome Project was just the beginning for understanding how genes affect the human body.
Study Finds 60 New Genes Controlled by DNA Snippet - URMC Press Room
