3/21/2006
Commercial genetic testing does not detect all cancer-associated mutations
Despite a negative (normal) genetic test for mutations in the BRCA1 and BRCA2 genes, about 12 percent of breast cancer patients from high-risk families carried previously undetected cancer-associated mutations, according to a study in the March 22/29 issue of JAMA, a theme issue on women's health.
Inherited mutations in BRCA1 and BRCA2 predispose to high risks of breast and ovarian cancer. Lifetime risks of breast cancer are as high as 80 percent among U.S. women with mutations in these genes, according to background information in the article. Risks for young women with inherited BRCA1 or BRCA2 mutations are particularly increased. Among white women in the U.S., 5 percent to 10 percent of breast cancer cases are due to inherited mutations in BRCA1 and BRCA2. Inherited mutations in other genes, including CHEK2, TP53 and PTEN, can also influence risk of breast cancer.
"The clinical dilemma is what to offer to women with a high probability of carrying a mutation in BRCA1 or BRCA2 but with negative commercial test results. Technically, the answer is at hand. The mutations identified in our study that were missed by commercial testing are detectable using other approaches that are currently available," the researchers write. They add that for families testing negative (wild type) for BRCA1 and BRCA2 by conventional sequencing, multiplex ligation-dependent probe amplification (MLPA - a molecular method to detect genetic variation) followed by sequence confirmation of breakpoints in patients' genomic DNA is the current best choice for evaluating the wide range of genomic rearrangements in BRCA1 and BRCA2. Clinical testing using MLPA is currently not available in the U.S.
"As more breast cancer susceptibility genes of different penetrances are identified, clinicians will be increasingly challenged to offer the most appropriate genetic tests, to assist patients in interpreting the results, and to optimize risk reduction strategies," the authors conclude. "Effective methods for identifying these mutations should be made available to women at high risk."
US commercial genetic testing does not detect all cancer-associated mutations in certain genes
Inherited mutations in BRCA1 and BRCA2 predispose to high risks of breast and ovarian cancer. Lifetime risks of breast cancer are as high as 80 percent among U.S. women with mutations in these genes, according to background information in the article. Risks for young women with inherited BRCA1 or BRCA2 mutations are particularly increased. Among white women in the U.S., 5 percent to 10 percent of breast cancer cases are due to inherited mutations in BRCA1 and BRCA2. Inherited mutations in other genes, including CHEK2, TP53 and PTEN, can also influence risk of breast cancer.
"The clinical dilemma is what to offer to women with a high probability of carrying a mutation in BRCA1 or BRCA2 but with negative commercial test results. Technically, the answer is at hand. The mutations identified in our study that were missed by commercial testing are detectable using other approaches that are currently available," the researchers write. They add that for families testing negative (wild type) for BRCA1 and BRCA2 by conventional sequencing, multiplex ligation-dependent probe amplification (MLPA - a molecular method to detect genetic variation) followed by sequence confirmation of breakpoints in patients' genomic DNA is the current best choice for evaluating the wide range of genomic rearrangements in BRCA1 and BRCA2. Clinical testing using MLPA is currently not available in the U.S.
"As more breast cancer susceptibility genes of different penetrances are identified, clinicians will be increasingly challenged to offer the most appropriate genetic tests, to assist patients in interpreting the results, and to optimize risk reduction strategies," the authors conclude. "Effective methods for identifying these mutations should be made available to women at high risk."
US commercial genetic testing does not detect all cancer-associated mutations in certain genes
The role of evolutionary genomics in the development of autism
Scientists at the London School of Economics, UK and Simon Fraser University, Canada have described the first hypothesis grounded in evolutionary genomics explaining the development of autism.
In an article to be published in a forthcoming issue of Journal of Evolutionary Biology, Dr Christopher Badcock and Professor Bernard Crespi explore the 'imprinted brain hypothesis' to explain the cause and effect of autism and autistic syndromes such as Asperger's syndrome, highlighted by the book The Curious Incident of the Dog in the Night-Time, which involves selective disruption of social behaviour that makes individuals more self-focussed whilst enhancing skills related to mechanistic cognition.
The 'imprinted brain hypothesis' suggests that competition between maternally and paternally expressed genes leads to conflicts within the autistic individual which could result in an imbalance in the brain's development. This is supported by the fact that there is known to be a strong genomic imprinting component to the genetic and developmental mechanisms of autism and autistic syndromes.
Professor Bernard Crespi from Simon Fraser University, Canada explains: "The imprinted brain hypothesis underscores the viewpoint that the autism spectrum represents human cognitive diversity rather than simply disorder or disability. Indeed, individuals at the highest-functioning end of this spectrum may have driven the development of science, engineering and the arts through mechanistic brilliance coupled with perseverant obsession."
The core behavioural features of autism such as self-focussed behaviour, altered social interactions and language and enhanced spatial and mechanistic cognition and abilities – as well as the degree to which the brain functions and structures are altered – also supports this hypothesis.
The role of evolutionary genomics in the development of autism
Full article:http://www.blackwell-synergy.com/doi/full/10.1111/j.1420-9101.2006.01091.x
In an article to be published in a forthcoming issue of Journal of Evolutionary Biology, Dr Christopher Badcock and Professor Bernard Crespi explore the 'imprinted brain hypothesis' to explain the cause and effect of autism and autistic syndromes such as Asperger's syndrome, highlighted by the book The Curious Incident of the Dog in the Night-Time, which involves selective disruption of social behaviour that makes individuals more self-focussed whilst enhancing skills related to mechanistic cognition.
The 'imprinted brain hypothesis' suggests that competition between maternally and paternally expressed genes leads to conflicts within the autistic individual which could result in an imbalance in the brain's development. This is supported by the fact that there is known to be a strong genomic imprinting component to the genetic and developmental mechanisms of autism and autistic syndromes.
Professor Bernard Crespi from Simon Fraser University, Canada explains: "The imprinted brain hypothesis underscores the viewpoint that the autism spectrum represents human cognitive diversity rather than simply disorder or disability. Indeed, individuals at the highest-functioning end of this spectrum may have driven the development of science, engineering and the arts through mechanistic brilliance coupled with perseverant obsession."
The core behavioural features of autism such as self-focussed behaviour, altered social interactions and language and enhanced spatial and mechanistic cognition and abilities – as well as the degree to which the brain functions and structures are altered – also supports this hypothesis.
The role of evolutionary genomics in the development of autism
Full article:http://www.blackwell-synergy.com/doi/full/10.1111/j.1420-9101.2006.01091.x
Researchers Find Fat Gene
NEW BRUNSWICK/PISCATAWAY, N.J. – Rutgers researchers have identified a gene – and the molecular function of its protein product – that provides an important clue to further understanding obesity and may point the way to new drugs to control fat metabolism.
The scientists found that the human protein known as lipin is a key fat-regulating enzyme. “Lipin activity may be an important pharmaceutical target for the control of body fat in humans, treating conditions that range from obesity to the loss of fat beneath the skin, as seen in HIV patients, ” said George M. Carman, a professor in Rutgers’ department of food science.
Rutgers Media Relations - Rutgers Researchers Find Fat Gene
The scientists found that the human protein known as lipin is a key fat-regulating enzyme. “Lipin activity may be an important pharmaceutical target for the control of body fat in humans, treating conditions that range from obesity to the loss of fat beneath the skin, as seen in HIV patients, ” said George M. Carman, a professor in Rutgers’ department of food science.
Rutgers Media Relations - Rutgers Researchers Find Fat Gene
Aggression-Related Gene Weakens Brain’s Impulse Control Circuits
A version of a gene previously linked to impulsive violence appears to weaken brain circuits that regulate impulses, emotional memory and thinking in humans, researchers at the National Institutes of Health's (NIH) National Institute of Mental Health (NIMH) have found. Brain scans revealed that people with this version — especially males — tended to have relatively smaller emotion-related brain structures, a hyperactive alarm center and under-active impulse control circuitry. The study identifies neural mechanisms by which this gene likely contributes to risk for violent and impulsive behavior through effects on the developing brain.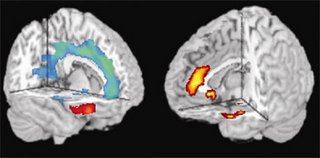
"These new findings illustrate the breathtaking power of 'imaging genomics' to study the brain's workings in a way that helps us to understand the circuitry underlying diversity in human temperament," said NIH Director Elias A. Zerhouni, M.D., who conducted MRI studies earlier in his career.
"By itself, this gene is likely to contribute only a small amount of risk in interaction with other genetic and psychosocial influences; it won't make people violent," explained Meyer-Lindenberg. "But by studying its effects in a large sample of normal people, we were able to see how this gene variant biases the brain toward impulsive, aggressive behavior."
The gene is one of two common versions that code for the enzyme monoamine oxydase-A (MAO-A), which breaks down key mood-regulating chemical messengers, most notably serotonin. The previously identified violence-related, or L, version, contains a different number of repeating sequences in its genetic code than the other version (H), likely resulting in lower enzyme activity and hence higher levels of serotonin. These, in turn, influence how the brain gets wired during development. The variations may have more impact on males because they have only one copy of this X-chromosomal gene, while females have two copies, one of which will be of the H variant in most cases.
The weakened regulatory circuits in men with L are compounded by intrinsically weaker connections between the orbital frontal cortex and amygdala in all men, say the researchers.
NIMH: Aggression-Related Gene Weakens Brain’s Impulse Control Circuits

"These new findings illustrate the breathtaking power of 'imaging genomics' to study the brain's workings in a way that helps us to understand the circuitry underlying diversity in human temperament," said NIH Director Elias A. Zerhouni, M.D., who conducted MRI studies earlier in his career.
"By itself, this gene is likely to contribute only a small amount of risk in interaction with other genetic and psychosocial influences; it won't make people violent," explained Meyer-Lindenberg. "But by studying its effects in a large sample of normal people, we were able to see how this gene variant biases the brain toward impulsive, aggressive behavior."
The gene is one of two common versions that code for the enzyme monoamine oxydase-A (MAO-A), which breaks down key mood-regulating chemical messengers, most notably serotonin. The previously identified violence-related, or L, version, contains a different number of repeating sequences in its genetic code than the other version (H), likely resulting in lower enzyme activity and hence higher levels of serotonin. These, in turn, influence how the brain gets wired during development. The variations may have more impact on males because they have only one copy of this X-chromosomal gene, while females have two copies, one of which will be of the H variant in most cases.
The weakened regulatory circuits in men with L are compounded by intrinsically weaker connections between the orbital frontal cortex and amygdala in all men, say the researchers.
NIMH: Aggression-Related Gene Weakens Brain’s Impulse Control Circuits
3/19/2006
Caught Up in DNA's Growing Web
Opinion piece by Harlan Levy, a former assistant district attorney in Manhattan, is a lawyer and the author of "And the Blood Cried Out."
Fifteen years ago, as a Manhattan homicide prosecutor, I was an aggressive proponent of taking DNA from convicted murderers, rapists and other violent felons so we could catch them when they committed crimes again.
I still firmly believe in the power of DNA to catch the guilty and exonerate the innocent.
But for all this technology's promise, proposals by some to extend DNA databanks far beyond convicted felons, and even to the general population, go too far.
In the early 1990's, state legislatures did what many early proponents of DNA urged: they passed laws to take DNA from those convicted of murder, rape and other violent felonies. Then they enacted laws to take DNA from most convicted felons. Misdemeanor sex crimes were next, a logical, intelligent measure.
But the proposed next steps in DNA collection were more problematic. In 1998, New York City's police commissioner, Howard Safir, proposed that DNA be taken from all arrestees. And Gov. George Pataki has sought to take DNA from people convicted of any misdemeanor, without proof that such offenders are more likely than the general population to commit violent felonies or sex crimes (the kinds of offenses where DNA evidence is most useful).
And the buzz today among prosecutors, judges and defense lawyers is that proposals to take DNA from the entire population are next.
What, if anything, is wrong with this picture? DNA databanks do help apprehend dangerous criminals (and thereby prevent crime). But most people aren't violent criminals and never will be, so putting their DNA on file exposes them to risks that they otherwise wouldn't face. First, the people who collect and analyze DNA can make mistakes (witness the Houston Police Department Laboratory, whose slapdash DNA procedures led to at least one wrongful conviction). Second, people can be framed by the police, a rival or an angry spouse. Third, DNA is all about context; there may be innocent reasons for a person's DNA to be at a crime scene, but the police are not always so understanding.
Indeed, with a universal national DNA databank, innocent people may be embroiled in criminal investigations when their DNA (a single hair or spot of saliva on a drinking glass) appears in a public or private place where they had every right to be.
Even if we get past those objections (do you trust the government with your DNA on file?), the practical barriers to universal collection loom larger still. In a nation with no institutionalized national identification cards, photo files or fingerprinting, just imagine requiring all citizens and residents to report to the local registry for DNA collection.
DNA databases should expand, but some fundamental principles should guide their development: government should aim DNA collection at those most likely to commit the crimes DNA can solve (rape and murder); before expanding collection, it should focus on improving laboratories and testing samples from unsolved violent crimes sitting untested in storage closets or refrigerators; and it should recognize (as have some but not all of our courts) that it does not have an unlimited right to every person's DNA without some showing of special need.
Caught Up in DNA's Growing Web - New York Times
Fifteen years ago, as a Manhattan homicide prosecutor, I was an aggressive proponent of taking DNA from convicted murderers, rapists and other violent felons so we could catch them when they committed crimes again.
I still firmly believe in the power of DNA to catch the guilty and exonerate the innocent.
But for all this technology's promise, proposals by some to extend DNA databanks far beyond convicted felons, and even to the general population, go too far.
In the early 1990's, state legislatures did what many early proponents of DNA urged: they passed laws to take DNA from those convicted of murder, rape and other violent felonies. Then they enacted laws to take DNA from most convicted felons. Misdemeanor sex crimes were next, a logical, intelligent measure.
But the proposed next steps in DNA collection were more problematic. In 1998, New York City's police commissioner, Howard Safir, proposed that DNA be taken from all arrestees. And Gov. George Pataki has sought to take DNA from people convicted of any misdemeanor, without proof that such offenders are more likely than the general population to commit violent felonies or sex crimes (the kinds of offenses where DNA evidence is most useful).
And the buzz today among prosecutors, judges and defense lawyers is that proposals to take DNA from the entire population are next.
What, if anything, is wrong with this picture? DNA databanks do help apprehend dangerous criminals (and thereby prevent crime). But most people aren't violent criminals and never will be, so putting their DNA on file exposes them to risks that they otherwise wouldn't face. First, the people who collect and analyze DNA can make mistakes (witness the Houston Police Department Laboratory, whose slapdash DNA procedures led to at least one wrongful conviction). Second, people can be framed by the police, a rival or an angry spouse. Third, DNA is all about context; there may be innocent reasons for a person's DNA to be at a crime scene, but the police are not always so understanding.
Indeed, with a universal national DNA databank, innocent people may be embroiled in criminal investigations when their DNA (a single hair or spot of saliva on a drinking glass) appears in a public or private place where they had every right to be.
Even if we get past those objections (do you trust the government with your DNA on file?), the practical barriers to universal collection loom larger still. In a nation with no institutionalized national identification cards, photo files or fingerprinting, just imagine requiring all citizens and residents to report to the local registry for DNA collection.
DNA databases should expand, but some fundamental principles should guide their development: government should aim DNA collection at those most likely to commit the crimes DNA can solve (rape and murder); before expanding collection, it should focus on improving laboratories and testing samples from unsolved violent crimes sitting untested in storage closets or refrigerators; and it should recognize (as have some but not all of our courts) that it does not have an unlimited right to every person's DNA without some showing of special need.
Caught Up in DNA's Growing Web - New York Times
Same Genes May Underlie Alcohol and Nicotine Co-Abuse
Vulnerability to both alcohol and nicotine abuse may be influenced by the same genetic factor, according to a recent study supported by the National Institute on Alcohol Abuse and Alcoholism (NIAAA), part of the National Institutes of Health (NIH).
In the study, two genetically distinct kinds of rat – one an innately heavy-drinking strain bred to prefer alcohol (“P” rats), the other strain bred to not prefer alcohol (“NP” rats) -- learned to give themselves nicotine injections by pressing a lever. Researchers found that P rats took more than twice as much nicotine as NP rats. Their findings were reported recently in the Journal of Neuroscience.
Researchers have known for some time that people who smoke are more likely to drink alcohol than non-smokers. Similarly, smoking is three times more common in people with alcoholism than in the general population. Since previous studies have also determined that genetics plays an important role in both alcohol and nicotine addictions, researchers have hypothesized that the same gene or genes may influence the co-abuse of these substances.
Same Genes May Underlie Alcohol and Nicotine Co-Abuse
In the study, two genetically distinct kinds of rat – one an innately heavy-drinking strain bred to prefer alcohol (“P” rats), the other strain bred to not prefer alcohol (“NP” rats) -- learned to give themselves nicotine injections by pressing a lever. Researchers found that P rats took more than twice as much nicotine as NP rats. Their findings were reported recently in the Journal of Neuroscience.
Researchers have known for some time that people who smoke are more likely to drink alcohol than non-smokers. Similarly, smoking is three times more common in people with alcoholism than in the general population. Since previous studies have also determined that genetics plays an important role in both alcohol and nicotine addictions, researchers have hypothesized that the same gene or genes may influence the co-abuse of these substances.
Same Genes May Underlie Alcohol and Nicotine Co-Abuse
Troubling times for embryo gene tests
AT THE age of 4, Doreen Flynn's first daughter, Jordan, was diagnosed with Fanconi anaemia, a rare genetic blood disorder that leaves people underweight and with a 700-fold greater chance of developing cancer. It is unlikely that Jordan will live past her early twenties.
Flynn and her husband wanted to have more children, partly because a bone-marrow transplant from a healthy sibling would be Jordan's best shot at survival. They decided to undergo pre-implantation genetic diagnosis, an IVF technique designed to ensure only healthy embryos are implanted. A Detroit-based lab, Genesis Genetics Institute, isolated two apparently disease-free embryos that would be bone-marrow matches for Jordan and implanted them at a hospital in Atlanta in early 2003. Julia and Jorjia were born 34 weeks later. Both have Fanconi anaemia.
Fortunately, reports of such errors connected to clinical genetic testing are extremely rare. Mark Hughes, the founder and chief clinician at Genesis, says that since the earliest days of his company 17 years ago, there have been only nine such instances out of several thousand tests conducted. "There is going to be an error rate in any diagnostics, and it's especially tricky in this instance," he says, because genetic testing relies upon identifying a very short DNA sequence among the large volume of DNA that comprises the human genome.
However, last week another case highlighted the potential costs of a lab or hospital getting a genetic test wrong. In what many are calling a "wrongful birth lawsuit", the Ohio Supreme Court upheld the right of Helen and Richard Schirmer to sue their healthcare provider, the Children's Hospital Medical Center in Cincinnati, for returning the wrong results of a fetal genetic test to diagnose trisomy 22, a genetic defect that causes severe mental and physical retardation. Their son, Matthew, was born with the condition in 1997.
Meanwhile, the diversity and popularity of genetic testing is skyrocketing. A recent survey by the OECD group of industrialised countries showed that the number of genetic tests carried out in these countries increased from 875,000 in 2000 to 1.4 million in 2002. There are now more than 900 distinct genetic tests in the US, compared with about 300 in 2002 – and that doesn't include the burgeoning number of "non-medical" hometesting kits sold to families over the internet.
This expansion is being driven both by a rapid growth in the number of genes discovered and advances in the technologies available to detect them.
In 2000, the US Centers for Medicare and Medicaid Services launched a notice of intent to create rules ensuring the quality of genetic testing, but difficulties in regulating the system have led to long delays. The OECD is attempting to develop similar guidelines for its member countries, which include the US, UK and much of Europe.
Troubling times for embryo gene tests
Pre-print from March 18 New Scientist
Flynn and her husband wanted to have more children, partly because a bone-marrow transplant from a healthy sibling would be Jordan's best shot at survival. They decided to undergo pre-implantation genetic diagnosis, an IVF technique designed to ensure only healthy embryos are implanted. A Detroit-based lab, Genesis Genetics Institute, isolated two apparently disease-free embryos that would be bone-marrow matches for Jordan and implanted them at a hospital in Atlanta in early 2003. Julia and Jorjia were born 34 weeks later. Both have Fanconi anaemia.
Fortunately, reports of such errors connected to clinical genetic testing are extremely rare. Mark Hughes, the founder and chief clinician at Genesis, says that since the earliest days of his company 17 years ago, there have been only nine such instances out of several thousand tests conducted. "There is going to be an error rate in any diagnostics, and it's especially tricky in this instance," he says, because genetic testing relies upon identifying a very short DNA sequence among the large volume of DNA that comprises the human genome.
However, last week another case highlighted the potential costs of a lab or hospital getting a genetic test wrong. In what many are calling a "wrongful birth lawsuit", the Ohio Supreme Court upheld the right of Helen and Richard Schirmer to sue their healthcare provider, the Children's Hospital Medical Center in Cincinnati, for returning the wrong results of a fetal genetic test to diagnose trisomy 22, a genetic defect that causes severe mental and physical retardation. Their son, Matthew, was born with the condition in 1997.
Meanwhile, the diversity and popularity of genetic testing is skyrocketing. A recent survey by the OECD group of industrialised countries showed that the number of genetic tests carried out in these countries increased from 875,000 in 2000 to 1.4 million in 2002. There are now more than 900 distinct genetic tests in the US, compared with about 300 in 2002 – and that doesn't include the burgeoning number of "non-medical" hometesting kits sold to families over the internet.
This expansion is being driven both by a rapid growth in the number of genes discovered and advances in the technologies available to detect them.
In 2000, the US Centers for Medicare and Medicaid Services launched a notice of intent to create rules ensuring the quality of genetic testing, but difficulties in regulating the system have led to long delays. The OECD is attempting to develop similar guidelines for its member countries, which include the US, UK and much of Europe.
Troubling times for embryo gene tests
Pre-print from March 18 New Scientist
NHGRI Announces New Sequencing Targets
BETHESDA, Md., Wed., Mar. 15, 2006 - The National Human Genome Research Institute (NHGRI), one of the National Institutes of Health (NIH), today announced its latest round of sequencing targets, with an emphasis on enhancing the understanding of how human genes function and how genomic differences between individuals influence the risk of health and disease.
The National Advisory Council for Human Genome Research, which is a federally chartered committee that advises NHGRI on program priorities and goals, recently approved three plans to specify the targets as part of its comprehensive strategy for NHGRI's Large-Scale Sequencing Research Network.
The plan given the highest priority is a project to identify structural variations in the human genome, which will characterize the most common types of structural variation in human DNA. The effort will use 48 human DNA samples donated for the recently completed International HapMap Project, which produced a comprehensive catalog of human genetic variation, or haplotypes, designed to speed the search for genes involved in common diseases.
The HapMap identified neighborhoods of tiny changes in DNA - known as single nucleotide polymorphisms (SNPs) - that can be involved in human disease. The structural variation effort will seek to identify instances where larger segments of DNA have been deleted, duplicated or rearranged - all of which can cause disease by disrupting the structure and function of genes.
A recent analysis has shown that these large-scale structural variations are much more common than previously appreciated. In fact, the genomes of any two humans are thought to differ by several hundred insertions, deletions and inversions.
The second plan will add DNA sequence to existing draft sequences of a number of primate species and add additional sequence information in regions of high biological interest within those genomes.
The third plan includes sequencing the genomes of eight new mammals at low-density draft coverage, which will be generated by sequencing their genomes at two-fold coverage.
Such comparisons between mammalian genomes represent one of the most effective ways to pinpoint the roughly five percent of the 3-billion base pair human genome that is most obviously functional.
genome.gov | 2006 Release: NHGRI Announces New Sequencing Targets
The National Advisory Council for Human Genome Research, which is a federally chartered committee that advises NHGRI on program priorities and goals, recently approved three plans to specify the targets as part of its comprehensive strategy for NHGRI's Large-Scale Sequencing Research Network.
The plan given the highest priority is a project to identify structural variations in the human genome, which will characterize the most common types of structural variation in human DNA. The effort will use 48 human DNA samples donated for the recently completed International HapMap Project, which produced a comprehensive catalog of human genetic variation, or haplotypes, designed to speed the search for genes involved in common diseases.
The HapMap identified neighborhoods of tiny changes in DNA - known as single nucleotide polymorphisms (SNPs) - that can be involved in human disease. The structural variation effort will seek to identify instances where larger segments of DNA have been deleted, duplicated or rearranged - all of which can cause disease by disrupting the structure and function of genes.
A recent analysis has shown that these large-scale structural variations are much more common than previously appreciated. In fact, the genomes of any two humans are thought to differ by several hundred insertions, deletions and inversions.
The second plan will add DNA sequence to existing draft sequences of a number of primate species and add additional sequence information in regions of high biological interest within those genomes.
The third plan includes sequencing the genomes of eight new mammals at low-density draft coverage, which will be generated by sequencing their genomes at two-fold coverage.
Such comparisons between mammalian genomes represent one of the most effective ways to pinpoint the roughly five percent of the 3-billion base pair human genome that is most obviously functional.
genome.gov | 2006 Release: NHGRI Announces New Sequencing Targets
Gene Influences Antidepressant Response
Whether depressed patients will respond to an antidepressant depends, in part, on which version of a gene they inherit, a study led by scientists at the National Institutes of Health (NIH) has discovered. Having two copies of one version of a gene that codes for a component of the brain's mood-regulating system increased the odds of a favorable response to an antidepressant by up to 18 percent, compared to having two copies of the other, more common version.
Since the less common version was over 6 times more prevalent in white than in black patients the researchers suggest that the gene may help to explain racial differences in the outcome of antidepressant treatment.
Everyone inherits two copies of the serotonin 2A receptor gene, one from each parent. A tiny glitch in the gene's chemical sequence results in some people having an adenine (A) at the same point that other people have a guanine (G). So an individual can have gene types AA, AG or GG. Overall, the prevalence of the A version was 38 percent, compared to 62 percent for the G version in this sample. Fourteen percent had AA gene type, 43 percent AG and 43 percent GG. Since the site of variation is located in a stretch of genetic material with no known function, the researchers suspect that it may be just a marker for a still-undiscovered functional variation nearby in the gene.
Based on scores on a depression rating scale, close to 80 percent of patients who had AA responded to the antidepressant, compared to about 62 percent of those with GG. Thus, patients with the AA gene type were 16-18 percent more likely to benefit from the medication. Even patients with AG showed some increased benefit.
NIMH: Gene Influences Antidepressant Response
Since the less common version was over 6 times more prevalent in white than in black patients the researchers suggest that the gene may help to explain racial differences in the outcome of antidepressant treatment.
Everyone inherits two copies of the serotonin 2A receptor gene, one from each parent. A tiny glitch in the gene's chemical sequence results in some people having an adenine (A) at the same point that other people have a guanine (G). So an individual can have gene types AA, AG or GG. Overall, the prevalence of the A version was 38 percent, compared to 62 percent for the G version in this sample. Fourteen percent had AA gene type, 43 percent AG and 43 percent GG. Since the site of variation is located in a stretch of genetic material with no known function, the researchers suspect that it may be just a marker for a still-undiscovered functional variation nearby in the gene.
Based on scores on a depression rating scale, close to 80 percent of patients who had AA responded to the antidepressant, compared to about 62 percent of those with GG. Thus, patients with the AA gene type were 16-18 percent more likely to benefit from the medication. Even patients with AG showed some increased benefit.
NIMH: Gene Influences Antidepressant Response
Scientists Find Psoriasis Gene
University of Michigan scientists have found a common genetic variation in an immune system gene that makes people much more likely to develop psoriasis – a disfiguring inflammatory skin disease.
Named PSORS1 (SORE-ESS-1), for psoriasis susceptibility 1, the gene is the first genetic determinant of psoriasis to be definitively identified in a large clinical study. Its discovery could lead to new, more effective treatments for psoriasis without the risks and side-effects of current therapies.
Unlike diseases caused by a mutation in just one gene, psoriasis is what scientists call a multi-factorial disease. This means that people must inherit several disease-related genes, plus be exposed to one or more environmental triggers, in order to get psoriasis.
The PSORS1 gene is actually one of over 20 different varieties (scientists call them alleles) of a gene called HLA-C. “In terms of our grocery store analogy, think of PSORS1 as one of 20 ‘brands’ of HLA-C on the shelf,” study director James T. Elder, M.D., Ph.D., says.
UMHS Press Release: U-M scientists find psoriasis gene
Named PSORS1 (SORE-ESS-1), for psoriasis susceptibility 1, the gene is the first genetic determinant of psoriasis to be definitively identified in a large clinical study. Its discovery could lead to new, more effective treatments for psoriasis without the risks and side-effects of current therapies.
Unlike diseases caused by a mutation in just one gene, psoriasis is what scientists call a multi-factorial disease. This means that people must inherit several disease-related genes, plus be exposed to one or more environmental triggers, in order to get psoriasis.
The PSORS1 gene is actually one of over 20 different varieties (scientists call them alleles) of a gene called HLA-C. “In terms of our grocery store analogy, think of PSORS1 as one of 20 ‘brands’ of HLA-C on the shelf,” study director James T. Elder, M.D., Ph.D., says.
UMHS Press Release: U-M scientists find psoriasis gene